Nested polymerase chain reaction
Nested polymerase chain reaction (nested PCR) is a modification of polymerase chain reaction intended to reduce non-specific binding in products due to the amplification of unexpected primer binding sites.[1]
Polymerase chain reaction
Polymerase chain reaction itself is the process used to amplify DNA samples, via a temperature-mediated DNA polymerase. The products can be used for sequencing or analysis, and this process is a key part of many genetics research laboratories, along with uses in DNA fingerprinting for forensics and other human genetic cases. Conventional PCR requires primers complementary to the termini of the target DNA. The amount of product from the PCR increases with the number of temperature cycles that the reaction is subjected to. A commonly occurring problem is primers binding to incorrect regions of the DNA, giving unexpected products. This problem becomes more likely with an increased number of cycles of PCR.
Primers
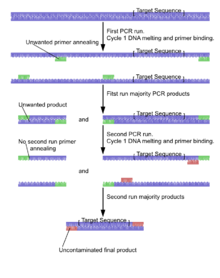
Nested polymerase chain reaction involves two sets of primers, used in two successive runs of polymerase chain reaction, the second set intended to amplify a secondary target within the first run product. This allows amplification for a low number of runs in the first round, limiting non-specific products. The second nested primer set should only amplify the intended product from the first round of amplification and not non-specific product. This allows running more total cycles while minimizing non-specific products. This is useful for rare templates or PCR with high background.
Processes
The target DNA undergoes the first run of polymerase chain reaction with the first set of primers, shown in green. The selection of alternative and similar primer binding sites gives a selection of products, only one containing the intended sequence.
The product from the first reaction undergoes a second run with the second set of primers, shown in red. It is very unlikely that any of the unwanted PCR products contain binding sites for both the new primers, ensuring the product from the second PCR has little contamination from unwanted products.
Text is available under the CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.
